Tutorial - Part #4 - Image Subtraction¶
Introduction¶
For image subtraction the package has a module called operations, which implements a main subtract function to estimate pair-image subtractions.
[1]:
from copy import copy
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors as colors
%matplotlib inline
[2]:
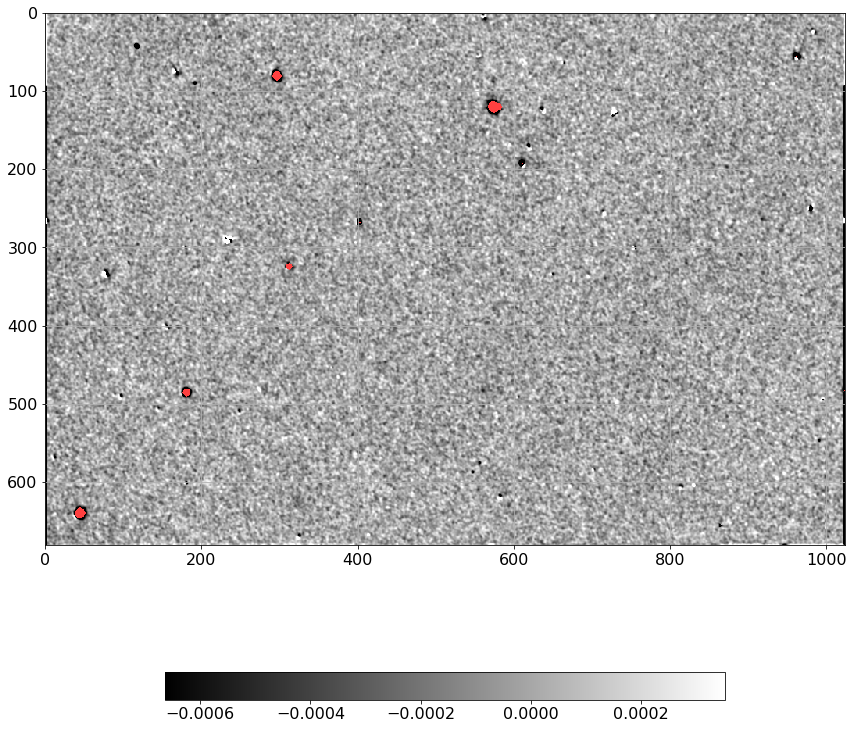
from astropy.io.fits import getdata
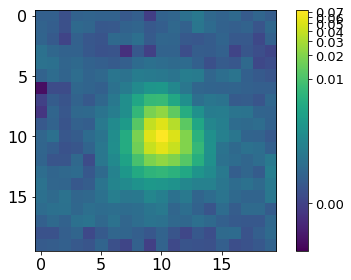
[3]:
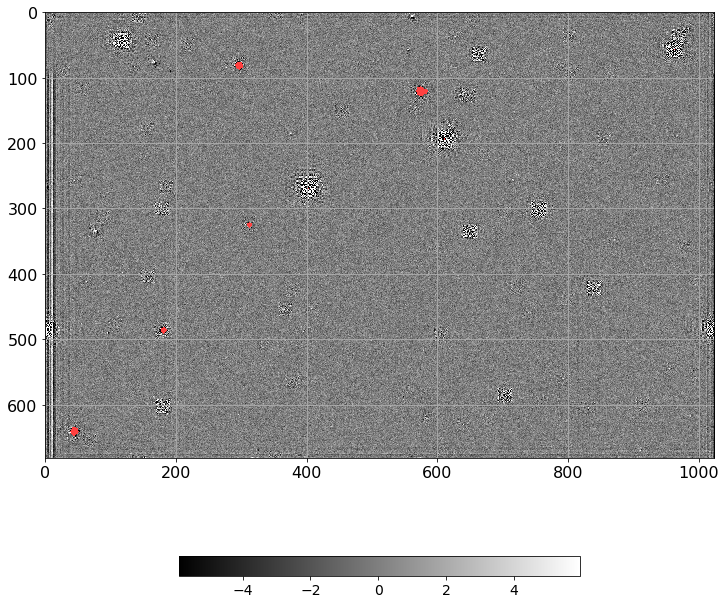
from astropy.visualization import LinearStretch, LogStretch
from astropy.visualization import ZScaleInterval, MinMaxInterval
from astropy.visualization import ImageNormalize
[4]:
palette = copy(plt.cm.gray)
palette.set_bad('r', 0.75)
[5]:
import properimage.single_image as si
from properimage.operations import subtract
[6]:
ref_path = './../../../data/aligned_eso085-030-004.fit'
new_path = './../../../data/aligned_eso085-030-005.fit'
To get the subtraction we need to run this function by using both paths for example:
[7]:
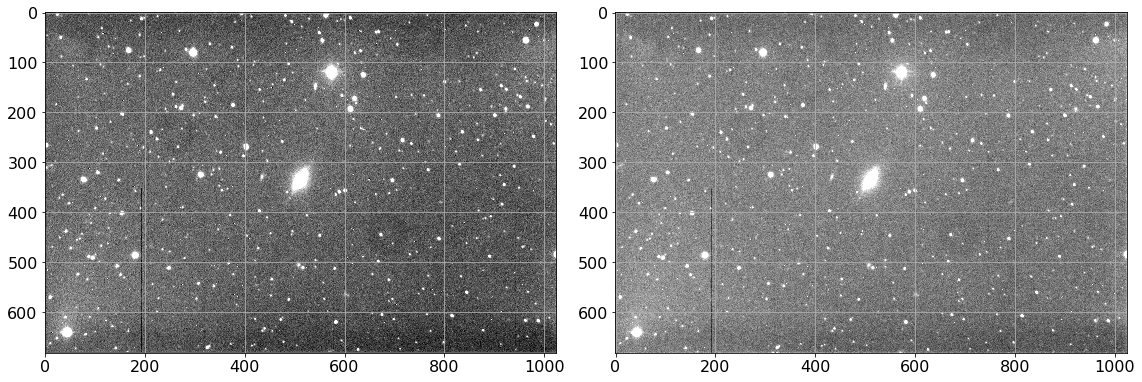
plt.figure(figsize=(16, 12))
plt.subplot(121)
ref = getdata(ref_path)
norm = ImageNormalize(ref, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.imshow(ref, cmap=plt.cm.gray, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.subplot(122)
ref = getdata(new_path)
norm = ImageNormalize(ref, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.imshow(ref, cmap=plt.cm.gray, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.tight_layout()
[8]:
%%time
result = subtract(
ref=ref_path,
new=new_path,
smooth_psf=False,
fitted_psf=True,
align=False,
iterative=False,
beta=False,
shift=False
)
updating stamp shape to (21,21)
updating stamp shape to (21,21)
CPU times: user 21 s, sys: 1.55 s, total: 22.6 s
Wall time: 18.2 s
The result is a list of numpy arrays.
The arrays are in order: D, P, Scorr, mask
[9]:
D = result[0]
P = result[1]
Scorr = result[2]
mask = result[3]
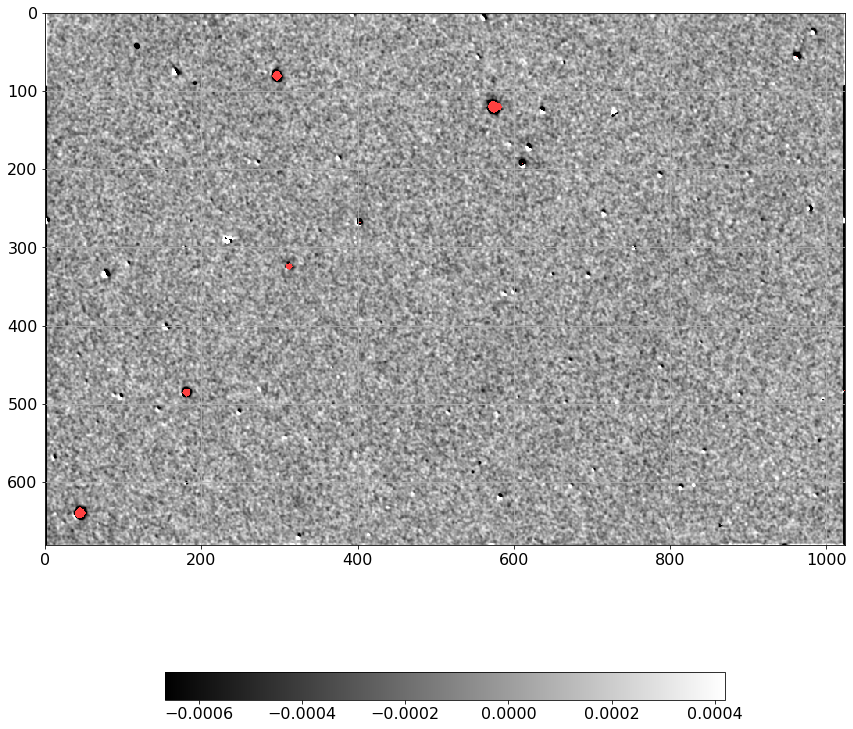
[10]:
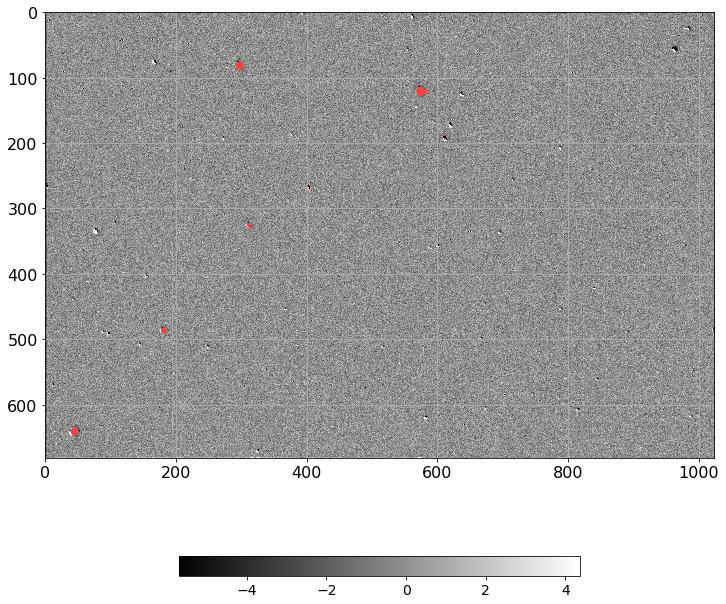
norm = ImageNormalize(D.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(D.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.colorbar(orientation='horizontal', shrink=0.6).ax.tick_params(labelsize=14)
[11]:
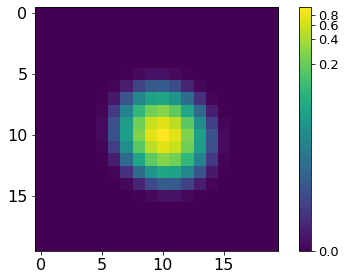
norm = ImageNormalize(P, interval=MinMaxInterval(), stretch=LogStretch())
xc, yc = np.where(P==P.max())
xc, yc = np.round(xc[0]), np.round(yc[0])
plt.imshow(P[xc-10:xc+10, yc-10:yc+10], norm=norm,
cmap='viridis', interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar().ax.tick_params(labelsize=13)
plt.tight_layout()
[12]:
norm = ImageNormalize(Scorr.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(Scorr.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar(orientation='horizontal', shrink=0.7).ax.tick_params(labelsize=16)
plt.grid()
plt.tight_layout()
[13]:
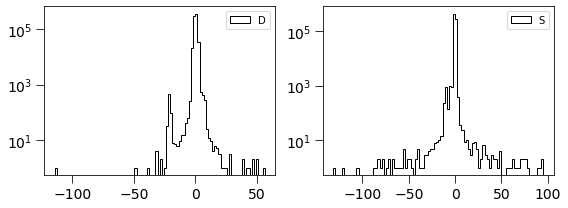
dimg = np.ma.MaskedArray(D.real, mask=mask).filled(0).flatten()
simage = np.ma.MaskedArray(Scorr.real, mask=mask).filled(0).flatten()
[14]:
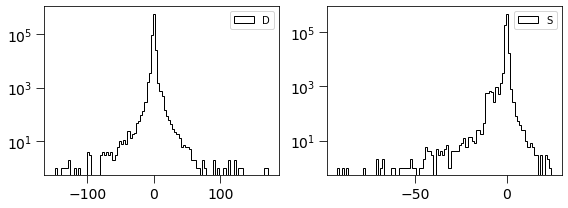
plt.figure(figsize=(8, 3))
plt.subplot(121)
plt.hist(dimg, log=True, bins=100, histtype='step', label='D', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.subplot(122)
plt.hist(simage/np.std(simage), log=True, bins=100, histtype='step', label='S', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.tight_layout()
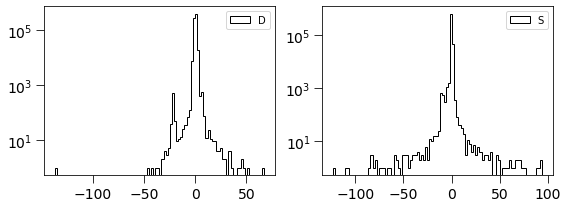
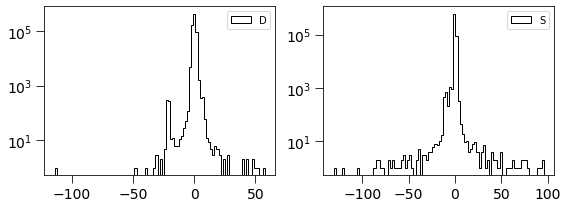
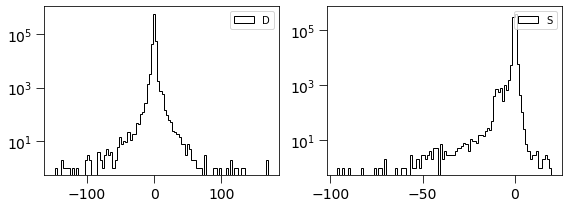
This is related to the quantities derived in Zackay et al. works. \(S_{corr} = P_D \otimes D\)
Different configurations:¶
We can run the subtraction using different configurations.
This parameters are the following:
align : bool
Whether to align the images before subtracting, default to False
inf_loss : float
Value of information loss in PSF estimation, lower limit is 0,
upper is 1. Only valid if fitted_psf=False. Default is 0.25
smooth_psf : bool
Whether to smooth the PSF, using a noise reduction technique.
Default to False.
beta : bool
Specify if using the relative flux scale estimation.
Default to True.
shift : bool
Whether to include a shift parameter in the iterative
methodology, in order to correct for misalignments.
Default to True.
iterative : bool
Specify if an iterative estimation of the subtraction relative
flux scale must be used. Default to False.
fitted_psf : bool
Whether to use a Gaussian fitted PSF. Overrides the use of
auto-psf determination. Default to True.
Example with beta=True, iterative=False¶
[15]:
%%time
D, P, Scorr, mask = subtract(
ref=ref_path,
new=new_path,
align=False,
iterative=False,
beta=True
)
updating stamp shape to (21,21)
updating stamp shape to (21,21)
CPU times: user 38.2 s, sys: 2.89 s, total: 41.1 s
Wall time: 37.5 s
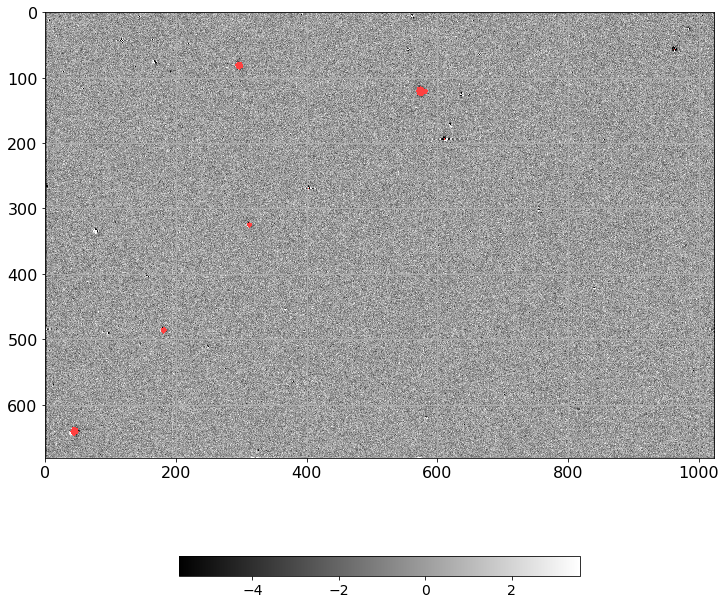
[16]:
norm = ImageNormalize(D.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(D.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.colorbar(orientation='horizontal', shrink=0.6).ax.tick_params(labelsize=14)
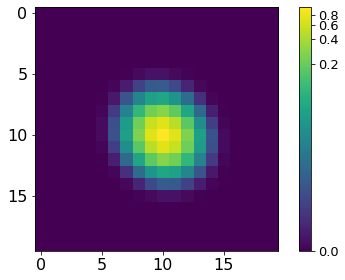
[17]:
norm = ImageNormalize(P, interval=MinMaxInterval(), stretch=LogStretch())
xc, yc = np.where(P==P.max())
xc, yc = np.round(xc[0]), np.round(yc[0])
plt.imshow(P[xc-10:xc+10, yc-10:yc+10], norm=norm,
cmap='viridis', interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar().ax.tick_params(labelsize=13)
plt.tight_layout()
[18]:
norm = ImageNormalize(Scorr.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(Scorr.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar(orientation='horizontal', shrink=0.7).ax.tick_params(labelsize=16)
plt.grid()
plt.tight_layout()
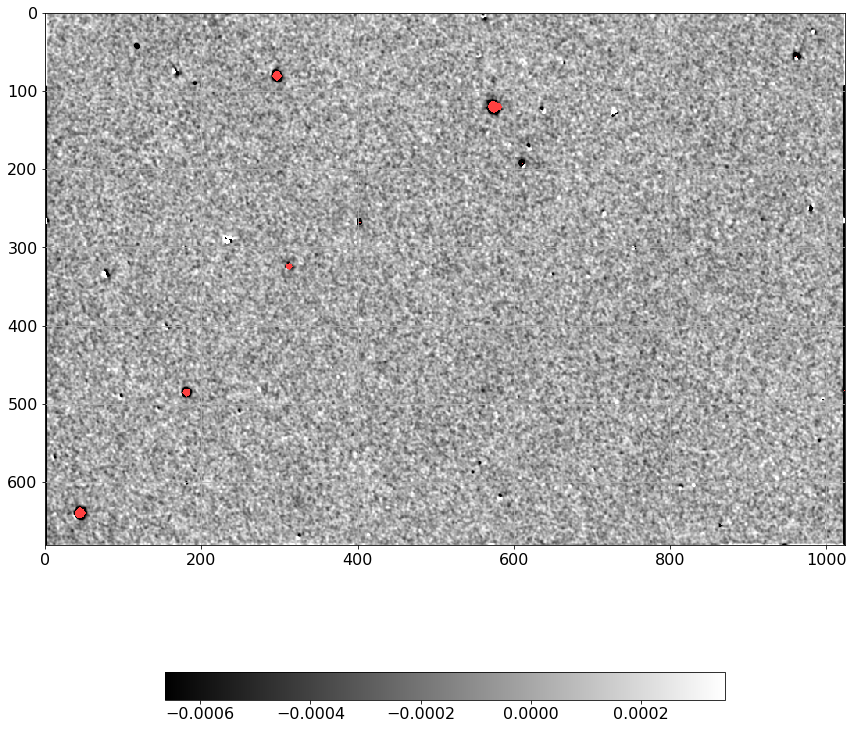
[19]:
dimg = np.ma.MaskedArray(D.real, mask=mask).filled(0).flatten()
simage = np.ma.MaskedArray(Scorr.real, mask=mask).filled(0).flatten()
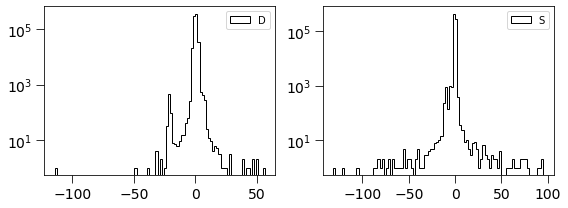
[20]:
plt.figure(figsize=(8, 3))
plt.subplot(121)
plt.hist(dimg, log=True, bins=100, histtype='step', label='D', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.subplot(122)
plt.hist(simage/np.std(simage), log=True, bins=100, histtype='step', label='S', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.tight_layout()
Example with iterative=True but beta=False¶
[21]:
%%time
D, P, Scorr, mask = subtract(
ref=ref_path,
new=new_path,
align=False,
iterative=True,
beta=False
)
updating stamp shape to (21,21)
updating stamp shape to (21,21)
CPU times: user 24.7 s, sys: 1.28 s, total: 25.9 s
Wall time: 22.6 s
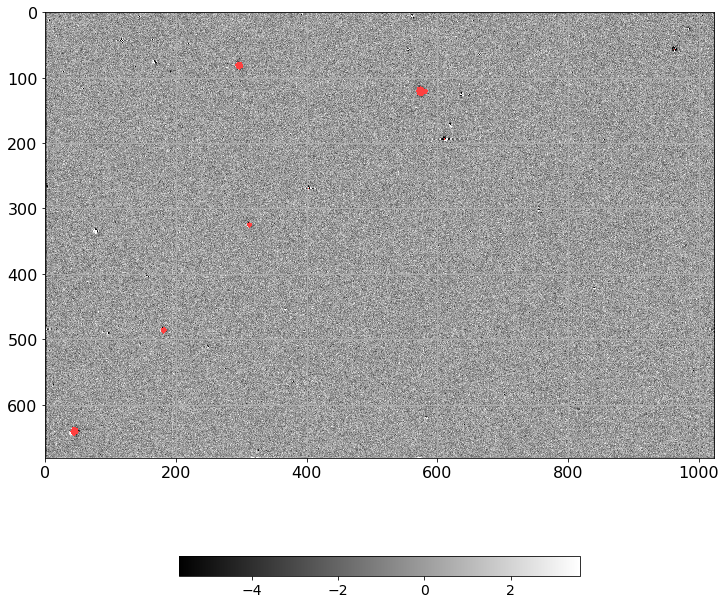
[22]:
norm = ImageNormalize(D.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(D.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.colorbar(orientation='horizontal', shrink=0.6).ax.tick_params(labelsize=14)
[23]:
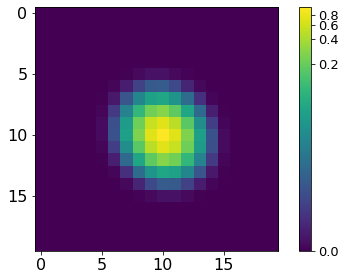
norm = ImageNormalize(P, interval=MinMaxInterval(), stretch=LogStretch())
xc, yc = np.where(P==P.max())
xc, yc = np.round(xc[0]), np.round(yc[0])
plt.imshow(P[xc-10:xc+10, yc-10:yc+10], norm=norm,
cmap='viridis', interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar().ax.tick_params(labelsize=13)
plt.tight_layout()
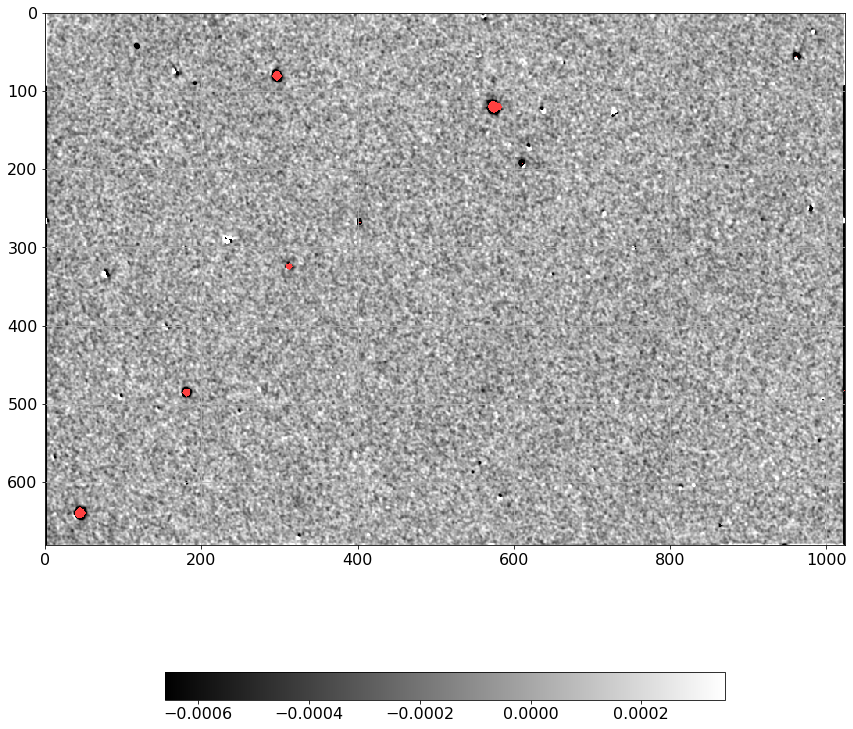
[24]:
norm = ImageNormalize(Scorr.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(Scorr.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar(orientation='horizontal', shrink=0.7).ax.tick_params(labelsize=16)
plt.grid()
plt.tight_layout()
[25]:
dimg = np.ma.MaskedArray(D.real, mask=mask).filled(0).flatten()
simage = np.ma.MaskedArray(Scorr.real, mask=mask).filled(0).flatten()
[26]:
plt.figure(figsize=(8, 3))
plt.subplot(121)
plt.hist(dimg, log=True, bins=100, histtype='step', label='D', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.subplot(122)
plt.hist(simage/np.std(simage), log=True, bins=100, histtype='step', label='S', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.tight_layout()
Example with iterative=True and beta=True¶
[27]:
%%time
D, P, Scorr, mask = subtract(
ref=ref_path,
new=new_path,
smooth_psf=False,
fitted_psf=True,
align=False,
iterative=True,
beta=True,
shift=True
)
updating stamp shape to (21,21)
updating stamp shape to (21,21)
CPU times: user 39 s, sys: 2.52 s, total: 41.5 s
Wall time: 38.2 s
The result is a list of numpy arrays.
The arrays are in order: D, P, Scorr, mask
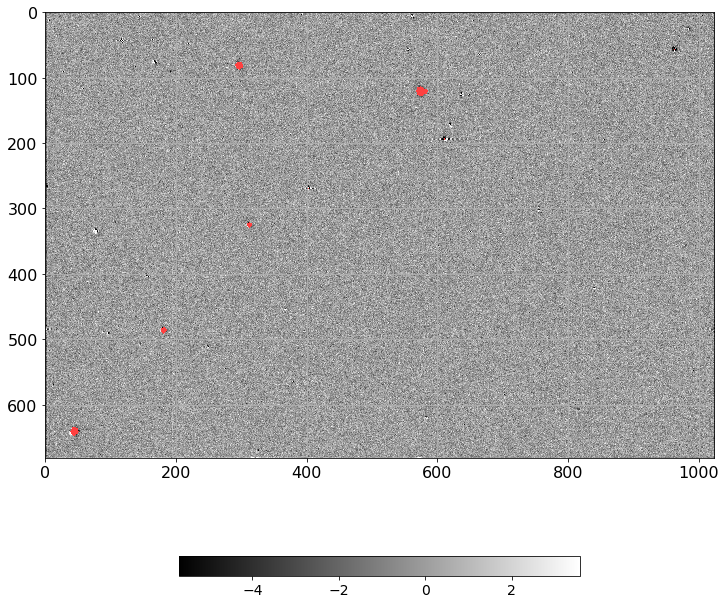
[28]:
norm = ImageNormalize(D.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(D.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.colorbar(orientation='horizontal', shrink=0.6).ax.tick_params(labelsize=14)
[29]:
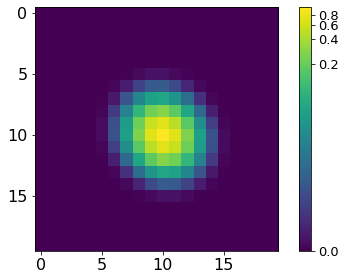
norm = ImageNormalize(P, interval=MinMaxInterval(), stretch=LogStretch())
xc, yc = np.where(P==P.max())
xc, yc = np.round(xc[0]), np.round(yc[0])
plt.imshow(P[xc-10:xc+10, yc-10:yc+10], norm=norm,
cmap='viridis', interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar().ax.tick_params(labelsize=13)
plt.tight_layout()
[30]:
norm = ImageNormalize(Scorr.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(Scorr.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar(orientation='horizontal', shrink=0.7).ax.tick_params(labelsize=16)
plt.grid()
plt.tight_layout()
[31]:
dimg = np.ma.MaskedArray(D.real, mask=mask).filled(0).flatten()
simage = np.ma.MaskedArray(Scorr.real, mask=mask).filled(0).flatten()
[32]:
plt.figure(figsize=(8, 3))
plt.subplot(121)
plt.hist(dimg, log=True, bins=100, histtype='step', label='D', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.subplot(122)
plt.hist(simage/np.std(simage), log=True, bins=100, histtype='step', label='S', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.tight_layout()
Example with fitted_psf=False and beta=True¶
[33]:
%%time
D, P, Scorr, mask = subtract(
ref=ref_path,
new=new_path,
smooth_psf=False,
fitted_psf=False,
align=False,
iterative=False,
beta=True,
shift=True
)
updating stamp shape to (21,21)
updating stamp shape to (21,21)
CPU times: user 37.1 s, sys: 1.48 s, total: 38.6 s
Wall time: 33.9 s
The result is a list of numpy arrays.
The arrays are in order: D, P, Scorr, mask
[34]:
norm = ImageNormalize(D.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(D.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.colorbar(orientation='horizontal', shrink=0.6).ax.tick_params(labelsize=14)
[35]:
norm = ImageNormalize(P, interval=MinMaxInterval(), stretch=LogStretch())
xc, yc = np.where(P==P.max())
xc, yc = np.round(xc[0]), np.round(yc[0])
plt.imshow(P[xc-10:xc+10, yc-10:yc+10], norm=norm,
cmap='viridis', interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar().ax.tick_params(labelsize=13)
plt.tight_layout()
[36]:
norm = ImageNormalize(Scorr.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(Scorr.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar(orientation='horizontal', shrink=0.7).ax.tick_params(labelsize=16)
plt.grid()
plt.tight_layout()
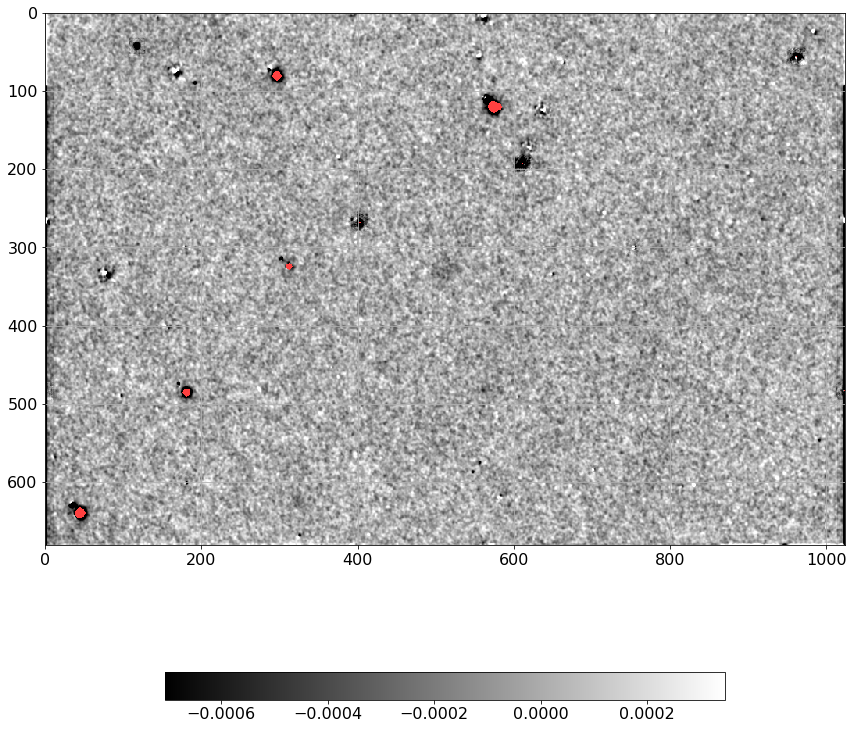
[37]:
dimg = np.ma.MaskedArray(D.real, mask=mask).filled(0).flatten()
simage = np.ma.MaskedArray(Scorr.real, mask=mask).filled(0).flatten()
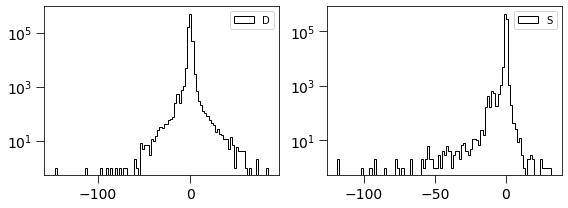
[38]:
plt.figure(figsize=(8, 3))
plt.subplot(121)
plt.hist(dimg, log=True, bins=100, histtype='step', label='D', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.subplot(122)
plt.hist(simage/np.std(simage), log=True, bins=100, histtype='step', label='S', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.tight_layout()
Example with smooth_psf=True and fitted_psf=False, using beta=True, iterative=True.¶
[39]:
%%time
D, P, Scorr, mask = subtract(
ref=ref_path,
new=new_path,
smooth_psf=True,
fitted_psf=False,
align=False,
iterative=True,
beta=True,
shift=True
)
updating stamp shape to (21,21)
updating stamp shape to (21,21)
CPU times: user 28 s, sys: 2.13 s, total: 30.1 s
Wall time: 25.8 s
The result is a list of numpy arrays.
The arrays are in order: D, P, Scorr, mask
[40]:
norm = ImageNormalize(D.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(D.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.colorbar(orientation='horizontal', shrink=0.6).ax.tick_params(labelsize=14)
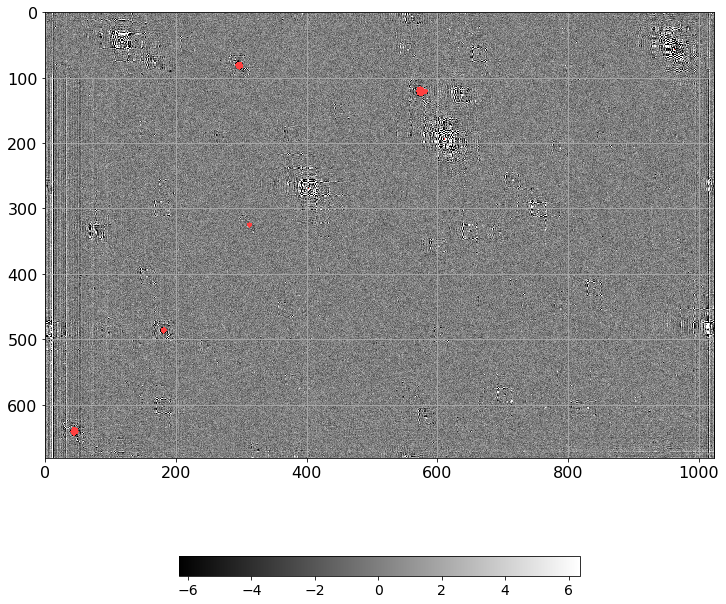
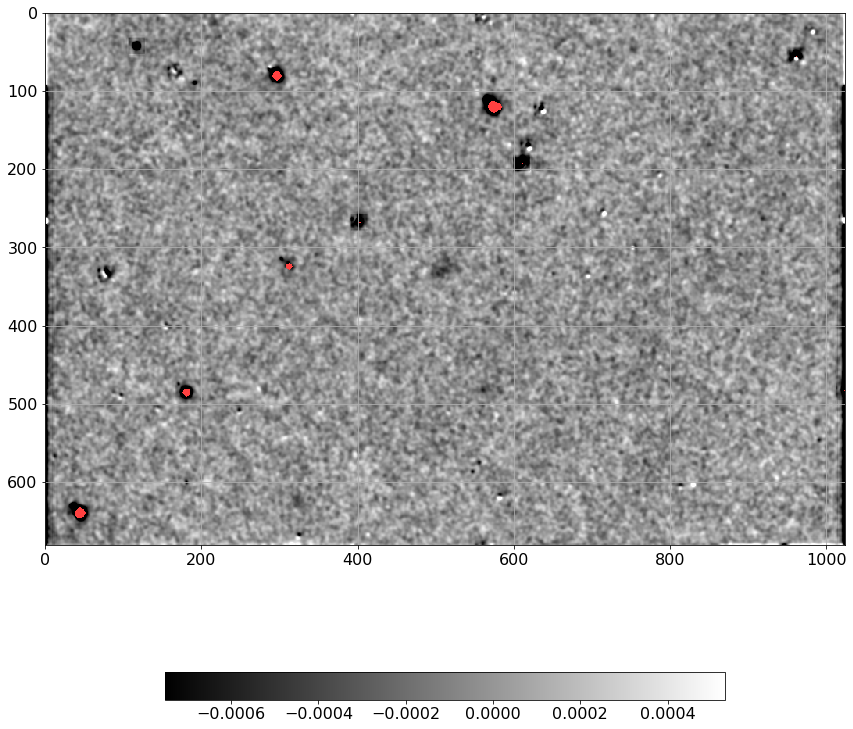
[41]:
norm = ImageNormalize(P, interval=MinMaxInterval(), stretch=LogStretch())
xc, yc = np.where(P==P.max())
xc, yc = np.round(xc[0]), np.round(yc[0])
plt.imshow(P[xc-13:xc+13, yc-13:yc+13], norm=norm,
cmap='viridis', interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar().ax.tick_params(labelsize=13)
plt.tight_layout()
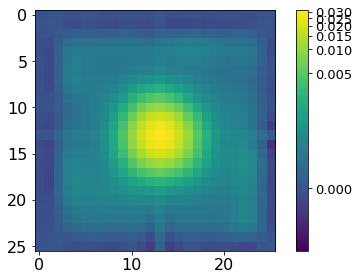
[42]:
norm = ImageNormalize(Scorr.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(Scorr.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar(orientation='horizontal', shrink=0.7).ax.tick_params(labelsize=16)
plt.grid()
plt.tight_layout()
[43]:
dimg = np.ma.MaskedArray(D.real, mask=mask).filled(0).flatten()
simage = np.ma.MaskedArray(Scorr.real, mask=mask).filled(0).flatten()
[44]:
plt.figure(figsize=(8, 3))
plt.subplot(121)
plt.hist(dimg, log=True, bins=100, histtype='step', label='D', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.subplot(122)
plt.hist(simage/np.std(simage), log=True, bins=100, histtype='step', label='S', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.tight_layout()
Example with smooth_psf=True and fitted_psf=False, using beta=False, iterative=False.¶
[45]:
%%time
D, P, Scorr, mask = subtract(
ref=ref_path,
new=new_path,
smooth_psf=True,
fitted_psf=False,
align=False,
iterative=False,
beta=False,
shift=True
)
updating stamp shape to (21,21)
updating stamp shape to (21,21)
CPU times: user 20.7 s, sys: 1.83 s, total: 22.5 s
Wall time: 17 s
The result is a list of numpy arrays.
The arrays are in order: D, P, Scorr, mask
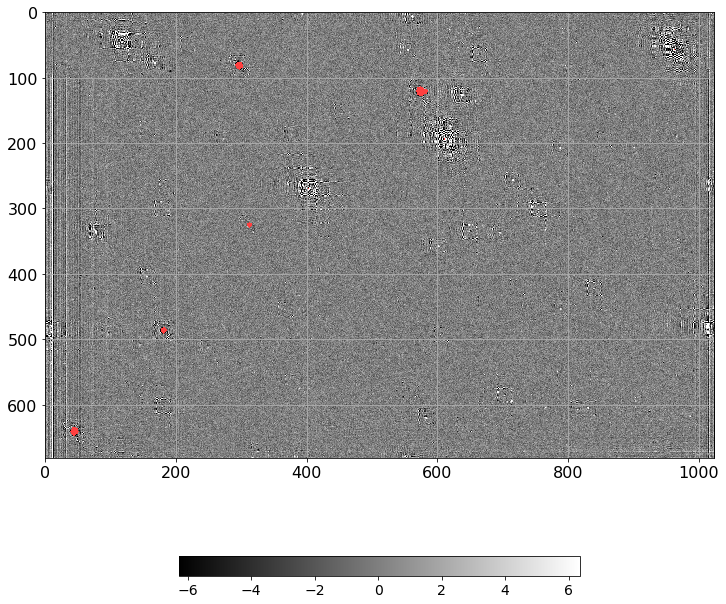
[46]:
norm = ImageNormalize(D.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(D.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.grid()
plt.colorbar(orientation='horizontal', shrink=0.6).ax.tick_params(labelsize=14)
[47]:
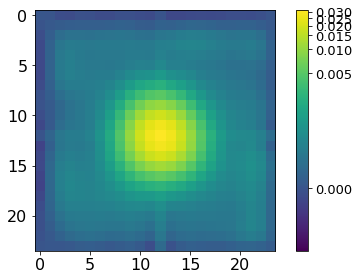
norm = ImageNormalize(P, interval=MinMaxInterval(), stretch=LogStretch())
xc, yc = np.where(P==P.max())
xc, yc = np.round(xc[0]), np.round(yc[0])
plt.imshow(P[xc-12:xc+12, yc-12:yc+12], norm=norm,
cmap='viridis', interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar().ax.tick_params(labelsize=13)
plt.tight_layout()
[48]:
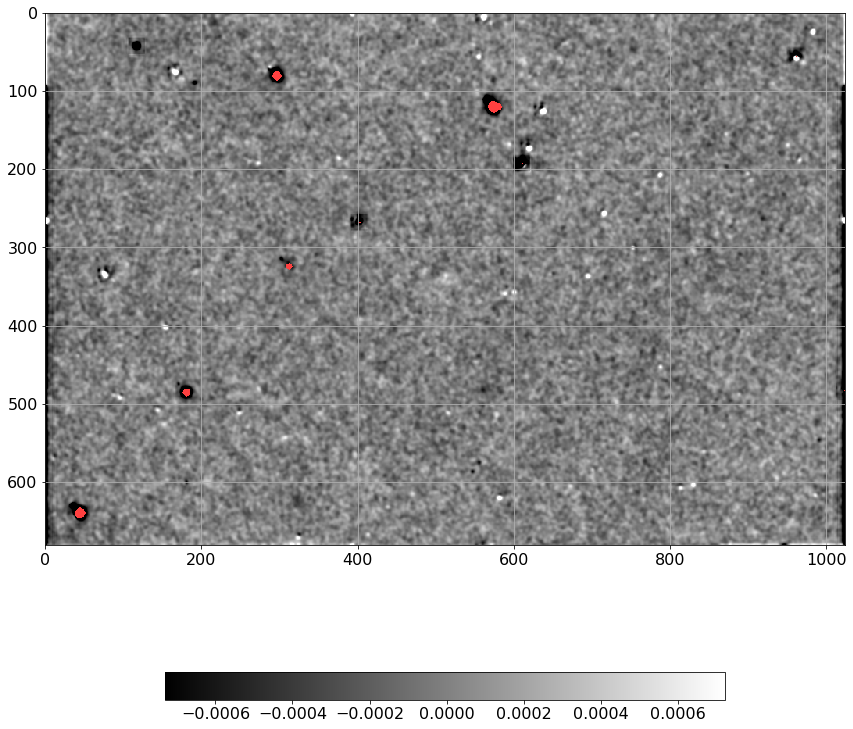
norm = ImageNormalize(Scorr.real, interval=ZScaleInterval(),
stretch=LinearStretch())
plt.figure(figsize=(12, 12))
plt.imshow(np.ma.MaskedArray(Scorr.real, mask=mask),
cmap=palette, norm=norm, interpolation='none')
plt.tick_params(labelsize=16)
plt.colorbar(orientation='horizontal', shrink=0.7).ax.tick_params(labelsize=16)
plt.grid()
plt.tight_layout()
[49]:
dimg = np.ma.MaskedArray(D.real, mask=mask).filled(0).flatten()
simage = np.ma.MaskedArray(Scorr.real, mask=mask).filled(0).flatten()
[50]:
plt.figure(figsize=(8, 3))
plt.subplot(121)
plt.hist(dimg, log=True, bins=100, histtype='step', label='D', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.subplot(122)
plt.hist(simage/np.std(simage), log=True, bins=100, histtype='step', label='S', color='k')
plt.legend(loc='best')
plt.tick_params(labelsize=14)
plt.tick_params(size=8)
plt.tight_layout()